![]() The CHPC has a new page summarizing machine learning and artifical intelligence resources.
The CHPC has a new page summarizing machine learning and artifical intelligence resources.
Center for High Performance Computing
Research Computing and Data Support for the University Community
In addition to deploying and operating high-performance computational resources and providing advanced user support and training, CHPC serves as an expert team to broadly support the increasingly diverse research computing and data needs on campus. These needs include support for big data, big data movement, data analytics, security, virtual machines, Windows science application servers, protected environments for data mining and analysis of protected health information, advanced networking, and more.
If you are new to the CHPC, the best place to learn about CHPC resources and policies is our Getting Started page.
Have a question? Please check our Frequently Asked Questions page and contact us if you require assistance or have further questions or concerns.

After nearly four decades of dedicated service at the University of Utah, Julia Harrison is retiring as the Operations Director of the Center for High Performance Computing.
Read more
Anita M. Orendt is a dedicated educator and researcher with a rich background in physical chemistry. Anita has made significant contributions to the academic community at the University of Utah.
Read moreUpcoming Events:
Allocation Requests for Winter 2025 are Due December 1st, 2024
Posted November 4th, 2024
Update to redwood idle session management following August 20, 2024 downtime
Posted September 3rd, 2024
Redwood Cluster Operating System Updated to Rocky Linux 8.10
Posted August 21st, 2024
Allocation Requests for Fall 2024 are Due September 1st, 2024
Posted August 7th, 2024
Allocation Requests for Summer 2024 are Due June 1st, 2024
Posted May 1st, 2024
CHPC Downtime: Tuesday March 5 starting at 7:30am
Posted February 8th, 2024
Two upcoming security related changes
Posted February 6th, 2024
Allocation Requests for Spring 2024 are Due March 1st, 2024
Posted February 1st, 2024
CHPC ANNOUNCEMENT: Change in top level home directory permission settings
Posted December 14th, 2023
CHPC Spring 2024 Presentation Schedule Now Available
CHPC PE DOWNTIME: Partial Protected Environment Downtime -- Oct 24-25, 2023
Posted October 18th, 2023
CHPC INFORMATION: MATLAB and Ansys updates
Posted September 22, 2023
CHPC SECURITY REMINDER
Posted September 8th, 2023
CHPC is reaching out to remind our users of their responsibility to understand what the software being used is doing, especially software that you download, install, or compile yourself. Read More...News History...
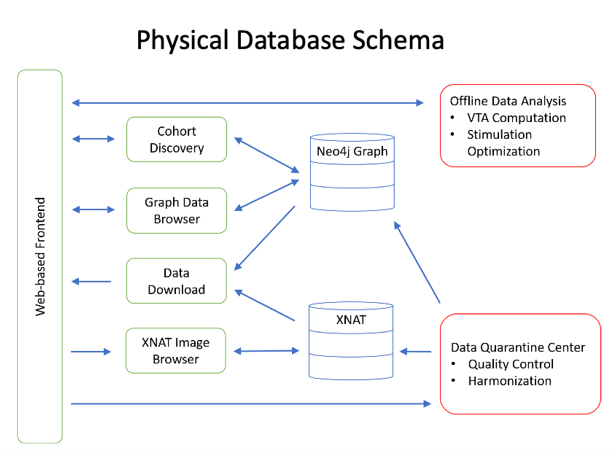
The International Neuromodulation Registry: A Graph Database Representation for Neuromodulation Therapies
By Hedges, D.M.1,2, Duffley, G.1,3, Hegman, J.C.1, Gouripeddi, R.2,4, Butson, C.R.1,3,5,6,7
1Scientific Computing and Imaging (SCI) Institute, 2Department of Biomedical Informatics, 3Department of Biomedical Engineering, 4Center for Clinical and Translational Science, 5Department of Neurology, 6Department of Neurosurgery, 7Department of Psychiatry, University of Utah
Deep Brain Stimulation (DBS) is a form of Neuromodulation therapy, often used in patients with many different types of neurological disorders. However, DBS is a rare treatment and medical centers have few patients who qualify for DBS, meaning that most DBS studies are statistically underpowered and have chronically low n values. Here, we present a platform designed to combine disparate datasets from different centers. Using this platform, researchers and clinicians will be able to aggregate patient datasets, transforming DBS studies from being center-based to being population-based.
Graph databases are increasing in popularity due to their speed of information retrieval, powerful visualization of complex data relationships, and flexible data models. Our Neo4j DBMS is physically located in the University of Utah Center for High-Performance Computing (CHPC) Protected Environment on a virtual machine, giving needs-based flexibility for both memory and storage.
This patient registry has been build on a next-generation graph database. Through a formal, but flexible, data model and ontology, this platform is able to harmonize disparate data types and allows for simple visualizations of complex data types.
Anticipated Use Cases: Cohort discovery, data and imaging download, exploratory analysis.
System Status
General Environment
| General Nodes | ||
|---|---|---|
| system | cores | % util. |
| kingspeak | 833/952 | 87.5% |
| notchpeak | 2545/3212 | 79.23% |
| lonepeak | 1585/1596 | 99.31% |
| Owner/Restricted Nodes | ||
| system | cores | % util. |
| ash | Status Unavailable | |
| notchpeak | 14288/21996 | 64.96% |
| kingspeak | 3550/5216 | 68.06% |
| lonepeak | 416/416 | 100% |
Protected Environment
| General Nodes | ||
|---|---|---|
| system | cores | % util. |
| redwood | 185/616 | 30.03% |
| Owner/Restricted Nodes | ||
| system | cores | % util. |
| redwood | 1690/6540 | 25.84% |